Berkeley Drosophila Genome Project
BDGP Resources
Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation
*For recovery of sequences flanking PZ, PlacW and PEP elements* *For SuPorP, pGT1, and p{Epgy2} elements, see the Bellen lab web site [\[HERE\]](http://flypush.imgen.bcm.tmc.edu/pscreen/)*
- Collect 30 anesthetized flies in eppendorf tube and freeze at -80°.
- Grind flies in 200 µl Buffer A with disposable tissue grinder (Kontes).
- Add an additional 200 µl Buffer A and continue grinding until only cuticles remain.
- Incubate at 65° for 30 minutes.
- Add 800 µl LiCl/KAc Solution and incubate on ice for at least 10 minutes.
- Spin for 15 minutes at RT.
- Transfer 1 ml of the supernatant into a new tube, avoiding floating crud.
- [If crud transfers, respin.]
- Add 600 µl isopropanol, mix, spin 15 minutes at RT.
- Aspirate away supernatant, pulse, aspirate, wash with 70% ethanol, dry.
- Resuspend in 150 µl TE.
- Store at -20°.
Buffer A
100 mM Tris-HCl, pH 7.5 100 mM EDTA 100 mM NaCl 0.5% SDS
_LiCl/KAc Solution _ Mix 1 part 5 M KAc stock : 2.5 parts 6 M LiCl stock
II. Digestions (Sau3A I, HinP1 I, or Msp I--done separately) Set up 2 separate digestions:
| Genomic DNA (~2 flies) | 10.0ul |
| 10X buffer (NEB Sau3A or NEB 2) | 2.5ul |
| 100 ug/ml RNase | 2.0ul |
| ddH2O | 8.0 or 9.5ul |
| Sau3A I, HinP1 I, or Msp I | 10 units |
- Incubate 2.5hrs at 37°.
- Heat for 20min to 65°.
- [Optional: Check 10ul of each digest on a 0.8% agarose gel.]
III. Ligations
| digested genomic DNA (~1 fly) | 10.0ul |
| 10X ligation buffer (+ATP) | 40.0ul |
| ddH2O | 350.0ul |
| ligase (2 Weiss units) | 2.0ul |
- Incubate O/N at 4°.
- Ethanol precipitate on ice, 10min.
- Spin, aspirate, pulse, aspirate.
- Speed Vac briefly to dry pellet.
- Resuspend in 150ul TE (~1/150 fly per ul).
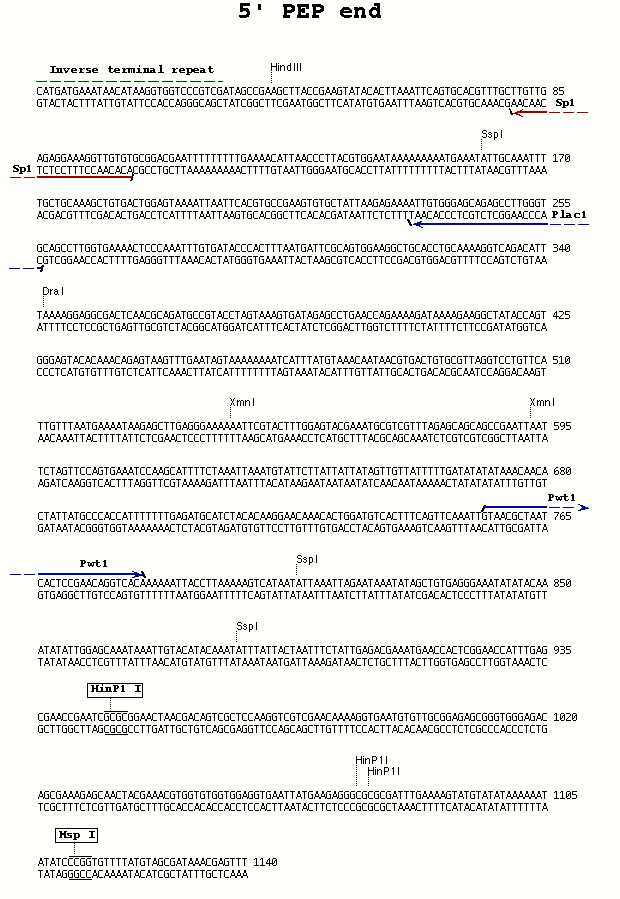
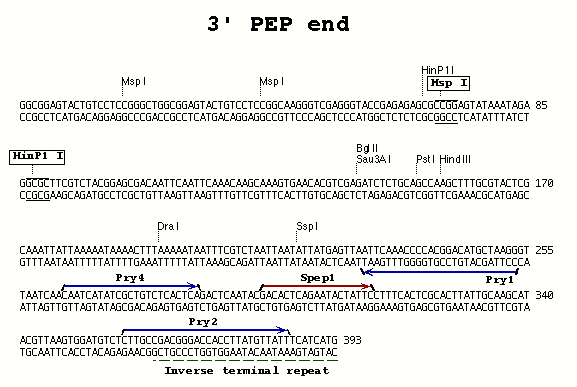
IV. PCR Set up PCR reactions with primers appropriate for the type of P element and the end of the element from which you want to recover genomic sequence (see table below and Figures).
| ligated genomic DNA (~1/15 fly) | 10.0ul |
| 2mM each dNTP | 2.0ul |
| 10uM forward primer | 1.0ul |
| 10uM reverse primer | 1.0ul |
| 10X Pharmacia Taq buffer | 5.0ul |
| ddH20 | 31.0ul |
| 2 units Taq | 0.4ul |
Primers for PCR:
| type of P element | 5' or 3' end | forward primer | reverse primer | annealing temperature |
|---|---|---|---|---|
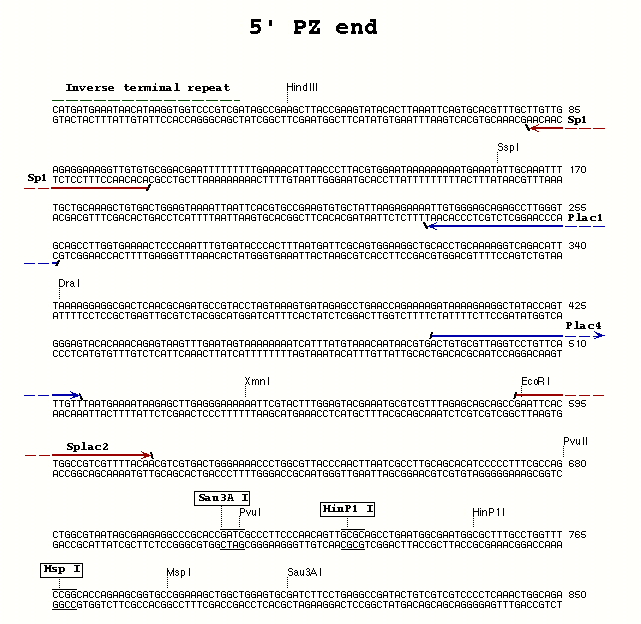
| PZ P-element | 5' end | Plac4 | Plac1 | 60° |
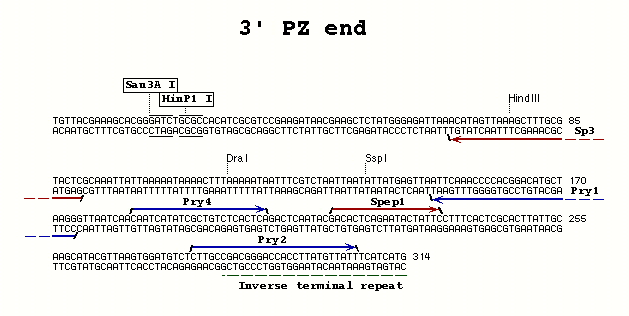
| PZ P-element | 3' end | Pry4 | Pry1 | 55° |
| PZ P-element | 3' end | Pry2 | Pry1 | 60° |
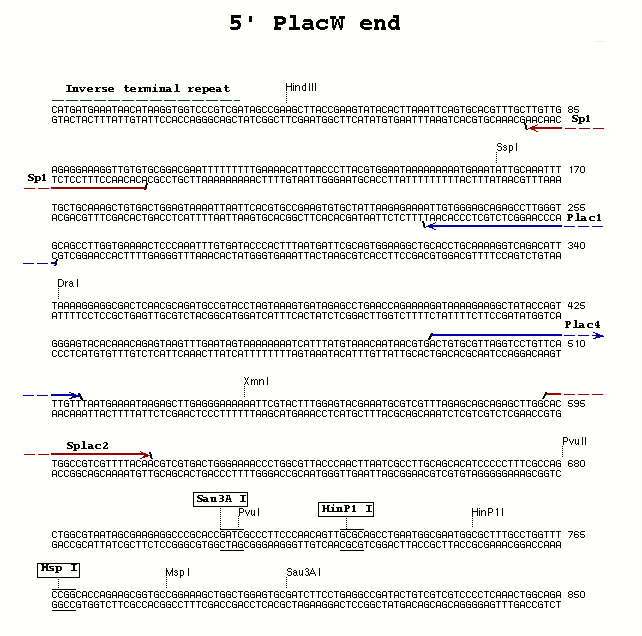
| PlacW P-element | 5' end | Plac4 | Plac1 | 60° |
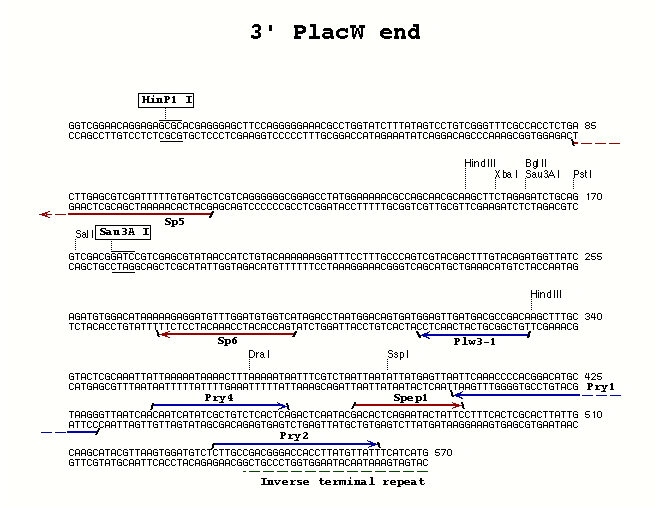
| PlacW P-element | 3' end | Pry4 | Plw3-1 | 55° |
| PlacW P-element | 3' end | Pry2 | Pry1 | 60° |
| PEP P-element | 5' end | Pwht1 | Plac1 | 60° |
| PEP P-element | 3' end | Pry4 | Pry1 | 55° |
| PEP P-element | 3' end | Pry2 | Pry1 | 60° |
The Pry2/Pry1 combination has a higher annealing temperature than the Pry4/Pry1 and Pry4/Plw3-1 combinations, but the resulting PCR products do not allow sequencing directly off the 3' end of the P-element. The latter primer combinations are therefore used in all initial experiments; the Pry2/Pry1 combination can be used in those cases where strong and unique bands do not result.
Cycle on MJ Research Thermal Cycler using the following parameters (~3hr run):
1X: 95° 5 min. 35X: 95° 30 sec./(see above) 1 min./68° 2 min. 1X: 72° 10 min. 4° hold
Note that these parameters have been optimized for the MJ Research cycler; other thermal cyclers have different cycling profiles and adjustments may be necessary to achieve optimal results.
Examine 5 ul of the PCR reactions on 1.5% agarose gel.
PCR Primer Sequences (5' to 3')--also see [**Figures**][3]:
**Plac4** (27) - ACT GTG CGT TAG GTC CTG TTC ATT GTT
**Plac1** (24) - CAC CCA AGG CTC TGC TCC CAC AAT
**Pry4** (23) - CAA TCA TAT CGC TGT CTC ACT CA
**Pry1** (26) - CCT TAG CAT GTC CGT GGG GTT TGA AT
**Pry2** (28) - CTT GCC GAC GGG ACC ACC TTA TGT TAT T
**Plw3-1** (19) - TGT CGG CGT CAT CAA CTC C
**Pwht1** (29) - GTA ACG CTA ATC ACT CCG AAC AGG TCA CA
V. Sequencing Generally strong and unique bands result from the above PCR conditions, which can be directly sequenced without extensive purifications. Prior to sequencing, simply pass PCR reactions through Qiagen PCR purification columns (P/N 28104) to remove primers and nucleotides and elute in 50-80 ul of 10 mM Tris. This step is essential when dye terminator or standard S-35 chemistry is used for sequencing and helpful when dye primer chemistry is used; although in this latter case it may not be essential when the primers used for sequencing are internal to those used for PCR. Cycle Sequencing Protocol Cycle sequence using protocols appropriate for your sequencing method. The following cycle sequencing protocol is used by Garson Tsang of BDGP (garson@fruitfly.berkeley.edu). Currently, we are using ABI 377XL Automated Sequencers, FMC Bioproducts Long Ranger gel solution (P/N 50611), and the Perkin Elmer ABI BigDye Terminator Cycle Sequencing Ready Reaction Kit (P/N 4303154).
The use of other thermal cyclers and sequencing strategies will require different cycling parameters and sequencing protocols.
-
Mix the following in 200 ul PCR tubes:
ABI BigDye Mix 3.0 ul PCR Template 2.5 ul Primer (1 uM) 1.5 ul 5X ABI dilution buffer 1.5 ul ddH2O 6.5 ul -
Cycle Sequence in a Perkin-Elmer 9700 (~2.5 hours) 96° 10 sec./ 50° 5 sec./ 60° 4 min. cycle 25 times followed by 4° hold
-
To purify reactions add cold 70% EtOH, let stand 20 minutes at RT, centrifuge for 30 minutes, discard EtOH and dry
-
Resuspend in 2.5 ul of the following: 5 parts (Deionized formamide) to 1 part (25 mM EDTA; 50 mg/ml Blue Dextran)
-
Load 1.9 ul onto gel
Pharmacia ALF Sequencers have also been used for direct cycle sequencing of PCR products using dye primer technology. Good results were obtained using the inexpensive Silver Sequencing System (Promega P/N Q4131 - for DNA Sequencing reagents only). It should be noted that the buffer and dideoxy/deoxy nucleotide mixes that come with this kit are not proprietary and can be easily prepared from stock reagents. The ALF sequencers are apparently more sensitive to excessively strong signals therefore the amount of PCR product added to the cycle sequencing reactions may need to be adjusted, especially in the case of very small products, in order to obtain acceptable sequence reads.
Standard S-35 sequencing should work as well; however, cycle sequencing protocols should be used for the sequencing reactions.
Sequencing primers--also see Figures.
The following primer sets are designed to sequence both ends of PCR products recovered from PlacW and PZ strains:
- Splac2 and Sp1 - for use with the Plac4/Plac1 5' PCR primer combination with either PZ or PlacW P-elements; allows sequencing of both ends of the PCR fragment
- Spep1 and Sp3 - for use with the Pry4/Pry1 3' PCR primer combination with PZ P-elements; allows sequencing of both ends of the PCR fragment
- Spep1 and Sp6 - for use with the Pry4/Plw3-1 3' PCR primer combination with PlacW P-elements where Sau3a digestion was performed; allows sequencing of both ends of the PCR fragment
- Spep1 and Sp5 - for use with the Pry4/Plw3-1 3' PCR primer combination with where HinP1 digestion was performed; allows sequencing of both ends of the PCR fragment
The PCR products recovered from PEP strains are sequenced with only one primer:
-
Sp1- for use with the Pwht1/Plac1 5' PCR primer combination with the PEP element
-
Spep1- for use with the Pry4/Pry1 3' PCR primer combination with the PEP element
Primer Sequences (5' to 3')---also see Figures:
Splac2 (25) - GAA TTC ACT GGC CGT CGT TTT ACA A Sp1 (22) - ACA CAA CCT TTC CTC TCA ACA A Sp3 (24) - GAG TAC GCA AAG CTT TAA CTA TGT Sp6 (23) - TGA CCA CAT CCA AAC ATC CTC TT Sp5 (25) - GCA TCA CAA AAA TCG ACG CTC AAG T Spep1 (19) - GAC ACT CAG AAT ACT ATT C
Melting temperatures of sequencing primers:
| Splac2 | 60.1ˆ |
| Sp1 | 50.6ˆ |
| Sp3 | 49.3ˆ |
| Sp6 | 54.9ˆ |
| Sp5 | 60.3ˆ |
| Spep1 | 44.8ˆ |
Figures: