Berkeley Drosophila Genome Project
Gene Disruption Project
Experimental Strategy of the P Element Controlled Misexpression Project
A number of groups have observed that a promoter or enhancer carried within a P element can be used to induce the misexpression of genes at the site of insertion (see for example Rørth, 1996. Proc. Natl. Acad. Sci. USA. 93: 12418-12422; Hay et al. 1997. Proc. Nat. Acad. Sci. USA 94: 5195-5200). We have begun to use Rørth's EP element (Rørth, 1996) that utilizes the versatile GAL4/UAS system (Brand and Perrimon, 1993. Development 118: 401-415) for controlled misexpression in our P mutagenesis project. In collaboration with Rørth's and S. Cohen's laboratories we constructed a set of 2,300 independent insertions of this element and characterized the phenotypes produced by these lines with several GAL4 drivers. These results have been extremely encouraging (Rørth et al., 1998. Development 125:1049-1057). The simple dominant overexpression screens reported in this paper identified a number of known genes implicated in pattern formation as well as a larger number of unknown genes. Overexpression phenotypes can result from expression of a gene in the wrong place or in the wrong amount. For example, hedgehog and dpp were identified because of spatial misexpression, since localized expression is important for their normal function. patched was identified because overexpression reduces Hedgehog signaling. Inappropriate expression of random proteins can also perturb development for a variety of non-specific reasons, for example reduced cell viability or failure of cells to differentiate appropriately. One way to minimize such effects is to look for cases where overexpression of a gene results in suppression of a mutant phenotype. Artifactual phenotypes should be less of a concern in such suppression screens than in simple overexpression screens. Suppressor screens are expected to select for genes or specific gene functions that are normally important for the process under study because they seek to restore wild-type cellular or developmental functions. The suppressor screen reported in Rørth et al.(1998) supports these contentions.
Outline of the modular misexpression screen.
Target lines each contain a single EP target element at an unknown position in the genome. When mated with GAL4
flies, progeny will contain both elements as shown below. This allows GAL4 to bind its sites within the EP element
and thereby induce the EP promoter to transcribe the gene immediately adjacent to the element. 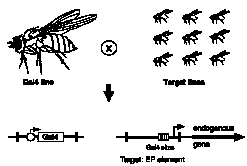
We are performing inverse PCR on these 2,300 lines to obtain a sequence tag that will precisely localize the insertion when the corresponding genomic region is sequenced. To date we have attempted this protocol on 891 EP lines. We obtained an average of 179 bp of flanking sequence; for 739 lines (83%) we obtained sufficient sequence (>25 bp) to provide a useful sequence tag. The 17% of the lines that failed in this initial procedure will be reanalyzed using a second set of PCR primers that amplify from the other end of the P element and/or by using an additional restriction enzyme. For inverse PCR to work unambiguously it is important that the lines contain only a single P element insertion. The Cytogenetics Core has mapped over 650 of these 2,300 lines to allow us to address this issue (and to provide additional mapped P elements on the X-chromosome where our other collections are deficient); over 97% of the lines examined contained a single P element insertion.
