Searches
ArmView: Instructions
Chromosome Arm Screen .. | .. STS Contig Screen .. | .. Help
The Chromosome Arm Screen
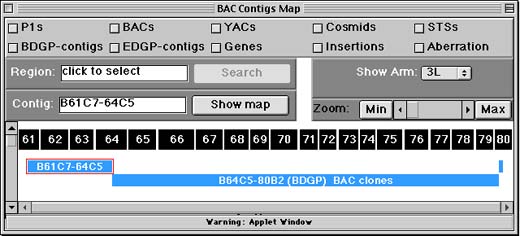
The Chromosome Arm Screen shows where Drosophila genome project contigs (contiguous, overlapping segments of BAC or P1 clones) overlap the classic cytological map. On the display, blue rectangles represent contigs now being sequenced as part of the Drosophila Genome Project. Green rectangles represent contigs for which no sequence data is yet available.
The black rectangles with numbers represent divisions, subdivisions, and bands in the salivary gland polytene chromosomes. Each rectangle is sized to match V. Sorsa's estimates for the nucleic acid content of each band - bigger rectangles represent bands Sorsa thought had more DNA than others. (See: Sorsa, V. 1988. Chromosome maps of Drosophila. CRC Press, Inc., Boca Raton, FL.)
How to: Show STS Content maps
To show a map for individual contigs,
- click the contig to select it.
- Next, click the "show map for contig" button - top left.
A new window with the STS map for the contig you chose should appear shortly. (The code for showing the map is loaded from our Web site -- it will take a few seconds.) This screen is explained in the STS Contig Screen section of this document.
How to: Search by region
- click items you want to retrieve (top of display).
options are: P1, YAC, BAC, and cosmid clones genomic DNA contigs (from EDGP or BDGP) genes, transposon insertions STSs, chromosomal aberrations
-
click chromosome bands to select a region (top display).
-
click "get checked items in" button (top left) to query Berkeley Fly Database
Your browser should open up a new window containing results for your search.
ArmView searches the Berkeley Fly Database. If you prefer a text-based query interface, the Berkeley Fly Database Query Form performs the same search as does ArmView.
And if you would like to use ArmView to search FlyBase, you can also use FlyBase ArmView.
The STS Contig Screen
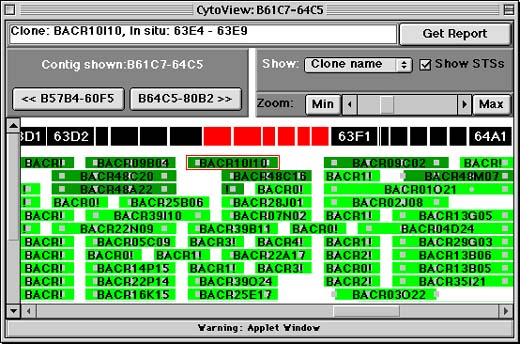
The STS Contig Screen displays a contiguous, overlapping set of BAC or P1 clones (a contig) as determined by the BDGP physical mapping project. BAC or P1 clones are shown as green horizontal rectangles, and Sequence Tag Sites (STSs) are displayed as gray squares, circles, and triangles (the shape corresponds to the source of the STS: squares for genomic clones, circles for genes, and triangles for transposons). If you click on either a clone or STS, ArmView will display its name and cytological position in the white text field at the top of the window. Clicking on the "Get Report" button allows you to get more information about the selected item.
The segmented black bar above the clones represents the cytological banding pattern of Drosophila's salivary polytene chromosomes. Bands are sized to match V. Sorsa's estimates for the nucleic acid content of each band - bigger rectangles represent bands Sorsa thought had more DNA than others. (See: Sorsa, V. 1988. Chromosome maps of Drosophila. CRC Press, Inc., Boca Raton, FL.) Because the contig is only roughly anchored to the cytological map, and because cytological readings are approximate, contig elements do not always appear directly below the band that actually contains them.
To scroll from right to left along the contig, use the scrollbar at the very bottom of the window. To zoom in or out to see more or less detail, use the "Zoom" controls on the right hand side of the window. The "Min" and "Max" buttons allow you to see the zoom in our out completely, while the scrollbar allows for more fine-grained control.
The << and >> buttons near the left hand side of the window allow you to skip directly to the adjacent contigs. Forward and backward are relative to the cytological division numbers in Drosophila.
Help
Printing the Map Viewer
To print the display, you must make a "screen shot" of the applet and save it to a file on your hard drive. This image file is also sometimes called a "screen capture" or "screen dump."
Next, use a graphics program (like Photoshop) to open the image file. Use the graphics program's "print" and "save" functions to print the image or save it in a different format.
If you can make screen captures and know how to use standard graphics software, then you should be able to cut and paste parts of the display into figures, slides, etc.
Mac and Windows computers come with screen capture support pre-installed. Follow the links below for details:
Comments and Questions About the Viewer Software
To contact us, send email to bdgp@fruitfly.berkeley.edu.
If you are reporting a problem, please tell us:
-
Name and version number of the browser you're using (e.g., Netscape Communicator 4.0 for Windows95)
-
A description of what went wrong (e.g., "couldn't select anything")
